Model Organism Databases and Bioinformatics
Our group co-directs the Xenopus model organism database, Xenbase. Xenopus is a type of African frog that is widely used in biomedical research. These frogs can be induced to lay eggs on demand, and it is straightforward to manipulate gene expression during their rapid development in a lab dish. Xenbase covers a wide variety of information types useful to people who use Xenopus in their research, everything from images of embryonic stages to the genomes of the two most commonly used species.

The database integrates developmental and genomic data on frog development from many different sources. Some content is gathered by data pipelines that query and update our content from remote sources, such as the NCBI, GenBank and other MODs. Our team of data curators also process scientific publications that used Xenopus and parse content from these papers into the database. Curators gather information such as what genes are examined, where are the genes expressed, what antibodies and other reagents were used, and how the experiments effect embryonic development.
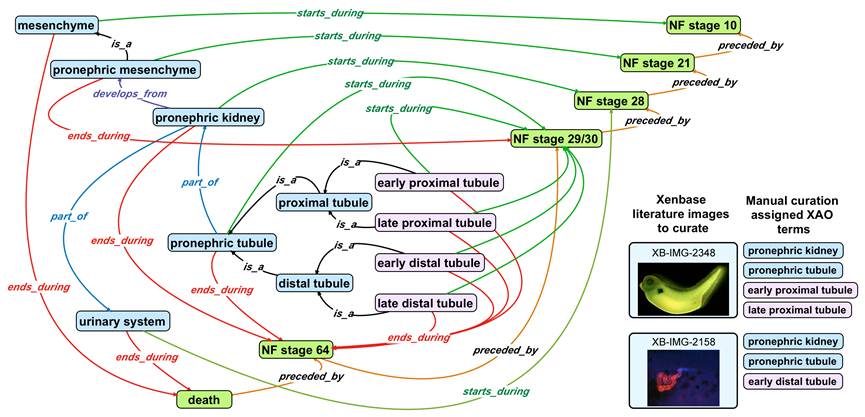
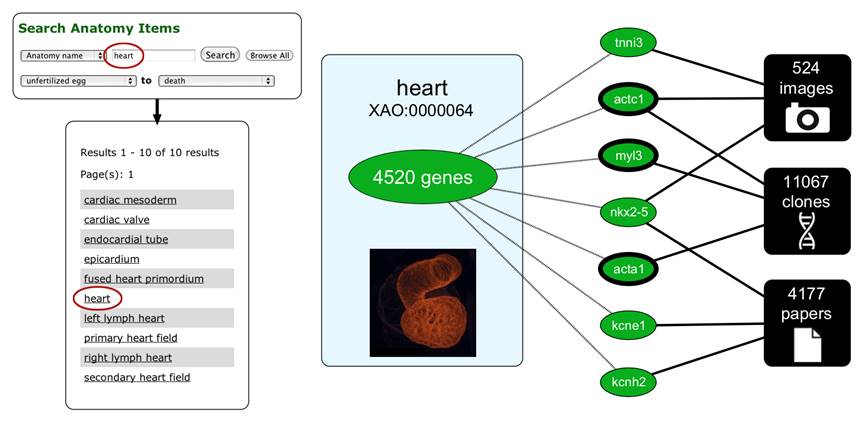
The Xenbase database schema integrates these different types of information using an ontology - The Xenopus Anatomical Ontology. A variety of different query interfaces allow visitors to search for gene expression patterns, reagents, available clones, promoter elements, etc.
Our research team runs the computational side of the project from server management, the database, the custom web application and the user interfaces. The project is funded by the NICHD under P41 HD064556
Visit the Xenopus database front end at xenbase.org
Key references
- Session et al. including Fortreide, J., Burns, K., Lotay, V., Karimi, K. and Vize, P.D. (2016). Genome evolution in the allotetraploid frog Xenopus laevis. Nature 538; 336-343
- Hellsten et al. including Vize, P.D. (2010) The genome of the western clawed frog Xenopus tropicalis. Science 328: 633-636.
- Vize, P.D. and Zorn, A.M. (2016) Xenopus genomic data and browser resources. Dev. Biol. pii: S0012-1606(15)30357-2. doi: 10.1016/j.ydbio.2016.03.030
- Vize, P.D. and Westerfield, M. (2015) Model Organism Databases. Genesis 53: 449.
- James-Zorn, C., Ponferrada, V.G., Burns, K., Fortreide, J., Lotay, V., Liu, Y., Karpinka, J.B., Karimi, K., Zorn, A.M. and Vize P.D. (2015). Xenbase; core features, data acquisition and data processing. Genesis 53: 486-497
- A.R.Deans et al. (inc. P.D. Vize), the Phenoscape Consortium. (2015) Finding our way through phenotypes. PLoS Biology, 13: e1002033.
- Karpinka, J.B., Fortreide, J.D., Burns, K.A., James-Zorn, C., Ponferrada, V.G., Lee, Jacqueline, Karimi, K., Zorn, A.M. and Vize, P.D. (2015). Xenbase, the Xenopus model organism database; new virtualized system, data types and genomes. Nucleic Acids Res. 43: D756-763
- Karimi, K. and Vize, P.D. (2014). The virtual Xenbase. Database 2014 ; doi: 10.1093/database/bau108
- Segerdell, E., Ponferrada V.G., James-Zorn C., Burns, K., Fortriede J., Dahdul W., Vize, P.D. and Zorn, A.M. (2013) XAO: an enhanced ontology of Xenopus anatomy and development. Journal of Biological Semantics 4:31 doi:10.1186/2041-1480-4-31